Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
Published:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Published:
This is a sample blog post. Lorem ipsum I can’t remember the rest of lorem ipsum and don’t have an internet connection right now. Testing testing testing this blog post. Blog posts are cool.
Short description of portfolio item number 1
Short description of portfolio item number 2 
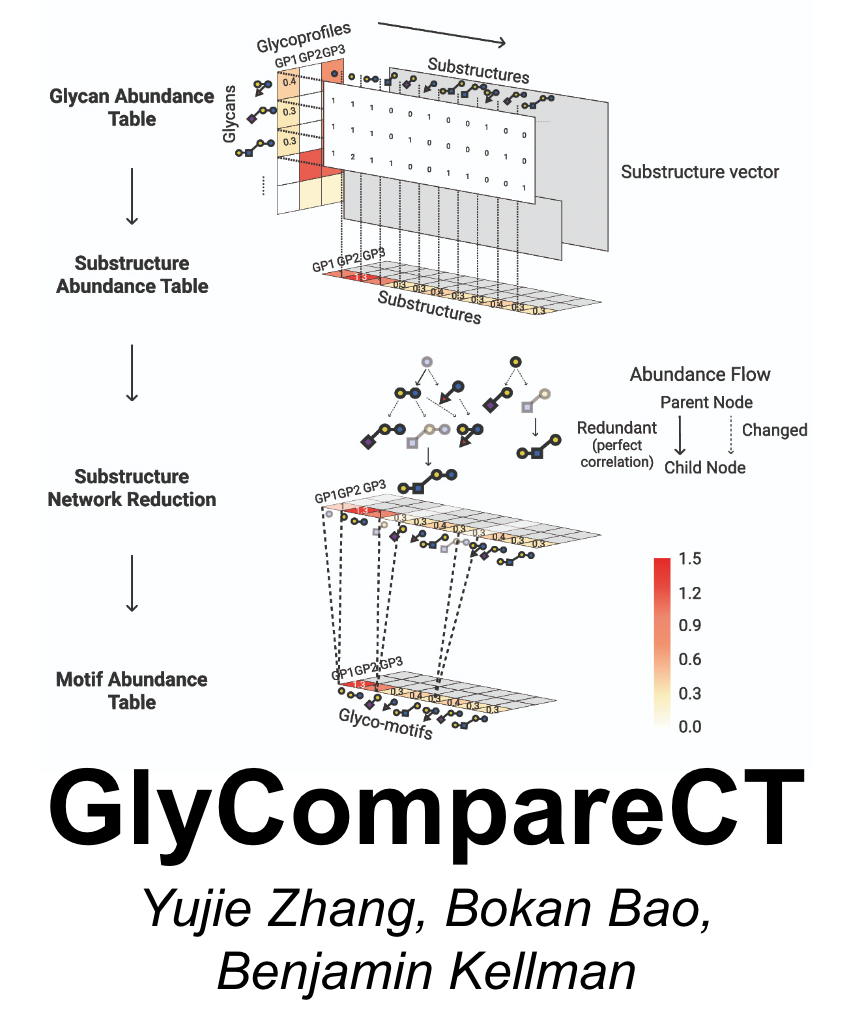
 GlyCompareCT is a portable command-line tool wrapping up the GlyCompare Python package. Given glycan abundances, GlyCompareCT conducts substructure decomposition to quantify hidden biosynthetic intermediate abundance and relationships between measured glycans. Thus, GlyComparCT mitigates sparsity and makes interdependence explicit, thereby increasing statistical power of downstream analyses of the glycomics data. Ultimately, GlyComparCT increases accessibility, interoperability, scope and consistency in glycomics analysis. It also largely saves the running memory than GlyCompare. GlyCompareCT GitHub
GlyCompareCT is a portable command-line tool wrapping up the GlyCompare Python package. Given glycan abundances, GlyCompareCT conducts substructure decomposition to quantify hidden biosynthetic intermediate abundance and relationships between measured glycans. Thus, GlyComparCT mitigates sparsity and makes interdependence explicit, thereby increasing statistical power of downstream analyses of the glycomics data. Ultimately, GlyComparCT increases accessibility, interoperability, scope and consistency in glycomics analysis. It also largely saves the running memory than GlyCompare. GlyCompareCT GitHub
 CardioDT is an imageJ-based program to detect and track cardiomyocytes contraction. The input is a microscopic video of cardiomyocytes contraction or other types of cells movements. The user will set threshold to detect cardiomyocytes as regions of interest (ROI) and then the program will calculate the ROI movement data and save it as excel files. It also provides a movement level separation function to cluster the cardiomyocytes with relative contraction degree. CardioDT GitHub
CardioDT is an imageJ-based program to detect and track cardiomyocytes contraction. The input is a microscopic video of cardiomyocytes contraction or other types of cells movements. The user will set threshold to detect cardiomyocytes as regions of interest (ROI) and then the program will calculate the ROI movement data and save it as excel files. It also provides a movement level separation function to cluster the cardiomyocytes with relative contraction degree. CardioDT GitHub

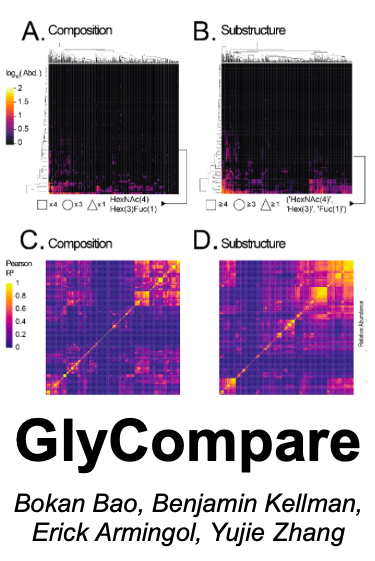 GlyCompare is a novel method wherein glycans from glycomic data are decomposed to a minimal set of intermediate substructures, thus incorporating shared intermediate glycan substructures into all comparisons of glycans. It used dynamic motif vectors to map new decomposed motifs thereby facilitating sample crossed comparisons. It inputs includes a glycan abundance table and a variable annotation file and outputs motif abundance table with optional clustering analysis graph. GlyCompare GitHub
GlyCompare is a novel method wherein glycans from glycomic data are decomposed to a minimal set of intermediate substructures, thus incorporating shared intermediate glycan substructures into all comparisons of glycans. It used dynamic motif vectors to map new decomposed motifs thereby facilitating sample crossed comparisons. It inputs includes a glycan abundance table and a variable annotation file and outputs motif abundance table with optional clustering analysis graph. GlyCompare GitHub
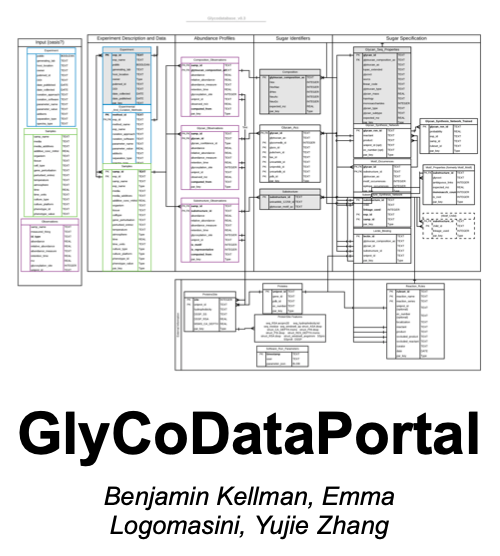
 GlyCoDataPortal is a database to facilitate glycan syntax transformation thereby speeds up the extraction of integrative insight. It bridges the gaps left by inconsistently identified glycans across datasets and tremendously enriching the information content of the data. It’s a private GitHub repo for lab use only.
GlyCoDataPortal is a database to facilitate glycan syntax transformation thereby speeds up the extraction of integrative insight. It bridges the gaps left by inconsistently identified glycans across datasets and tremendously enriching the information content of the data. It’s a private GitHub repo for lab use only.
Published in Preprint, 2020
This paper is about showing that commensal host bacterial communities can modify HS and thereby modulate SARS-CoV-2 spike protein binding and that these communities change with host age and sex.
Recommended citation: Martino, Cameron, et al. "Bacterial modification of the host glycosaminoglycan heparan sulfate modulates SARS-CoV-2 infectivity." bioRxiv (2020). https://doi.org/10.1101/2020.08.17.238444
Published in Beilstein Journal of Organic Chemistry, 2020
This paper is about an unambiguous representation for describing glycosylation reactions in both literature and code.
Recommended citation: Kellman, Benjamin P., Yujie Zhang, Emma Logomasini, Eric Meinhardt, Karla P. Godinez-Macias, Austin WT Chiang, James T. Sorrentino et al. "A consensus-based and readable extension of Linear Code for Reaction Rules (LiCoRR)." Beilstein journal of organic chemistry 16, no. 1 (2020): 2645-2662. https://doi.org/10.3762/bjoc.16.215
Published in Nature Communications, 2021
This paper is about a glycomic data analysis tool called glyCompare and its demonstration.
Recommended citation: Bao, Bokan, Benjamin P. Kellman, Austin WT Chiang, Yujie Zhang, James T. Sorrentino, Austin K. York, Mahmoud A. Mohammad, Morey W. Haymond, Lars Bode, and Nathan E. Lewis. "Correcting for sparsity and interdependence in glycomics by accounting for glycan biosynthesis." Nature communications 12, no. 1 (2021): 1-14. https://doi.org/10.1038/s41467-021-25183-5
Published in preprint, 2023
This paper is about a new tool for reconstructing satellite repeat units and HORs from accurate reads or assemblies without prior knowledge on repeat structures.
Recommended citation: Zhang, Yujie, Justin Chu, Haoyu Cheng, and Heng Li. "De novo reconstruction of satellite repeat units from sequence data." arXiv preprint arXiv:2304.09729 (2023). https://doi.org/10.48550/arXiv.2304.09729
Published in STAR protocols, 2023
This paper is a protocol of using GlyCompareCT, a command-line glycomic data analysis tool based on GlyCompare.
Recommended citation: Zhang, Yujie, Sridevi Krishnan, Bokan Bao, Austin WT Chiang, James T. Sorrentino, Song-Min Schinn, Benjamin P. Kellman, and Nathan E. Lewis. "Preparing glycomics data for robust statistical analysis with GlyCompareCT." STAR protocols 4, no. 2 (2023): 102162. https://doi.org/10.1016/j.xpro.2023.102162
Published:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Published:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.
Undergraduate course, University 1, Department, 2014
This is a description of a teaching experience. You can use markdown like any other post.
Workshop, University 1, Department, 2015
This is a description of a teaching experience. You can use markdown like any other post.